GSP_DEMO_WAVELET - Introduction to spectral graph wavelet with the GSPBox
Description
The wavelets are a special type of filterbank. In this demo, we will show how you can very easily construct a wavelet frame and apply it to a signal. If you want to do find an interactive demo of the wavelet, we encourage you to use the sgwt_demo2 of the sgwt toolbox. It can be downloaded at:
http://wiki.epfl.ch/sgwt/documents/sgwt_toolbox-1.02.zip
The sgwt toolbox has the same core as the GSPBox and all his functions have equivalent in the GSPBox ( Except the demos ;-) ).
In this demo we will show you how to compute the wavelet coefficients of a graph and visualize them. First, let's load a graph
G = gsp_bunny();
This graph is a nearest-neighbor graph of a pointcloud of the Stanford bunny. It will allow us to get interesting visual results using wavelets.
At this stage we could compute the full Fourier basis using gsp_compute_fourier_basis, but this would take a lot of time, and can be avoided by using Chebychev polynomials approximations. This operation is implemented in most function and is thus completely transparent.
Simple filtering
Before tackling wavelets, we can see the effect of one filter localized on the graph. So we can first design a few heat kernel filters
taus = [1, 10, 25, 50]; Hk = gsp_design_heat(G, taus);
Let's now create a signal as a Kronecker located on one vertex (e.g. the vertex 100)
S = zeros(G.N, 1); vertex_delta = 83; S(vertex_delta) = 1; Sf_vec = gsp_filter_analysis(G, Hk, S); Sf = gsp_vec2mat(Sf_vec, length(taus));
and plot the filtered signal
param_plot.cp = [0.1223, -0.3828, 12.3666];
figure;
subplot(221)
gsp_plot_signal(G,Sf(:,1), param_plot);
axis square
title(sprintf('Heat diffusion tau = %d', taus(1)));
subplot(222)
gsp_plot_signal(G,Sf(:,2), param_plot);
axis square
title(sprintf('Heat diffusion tau = %d', taus(2)));
subplot(223)
gsp_plot_signal(G,Sf(:,3), param_plot);
axis square
title(sprintf('Heat diffusion tau = %d', taus(3)));
subplot(224)
gsp_plot_signal(G,Sf(:,4), param_plot);
axis square
title(sprintf('Heat diffusion tau = %d', taus(4)));
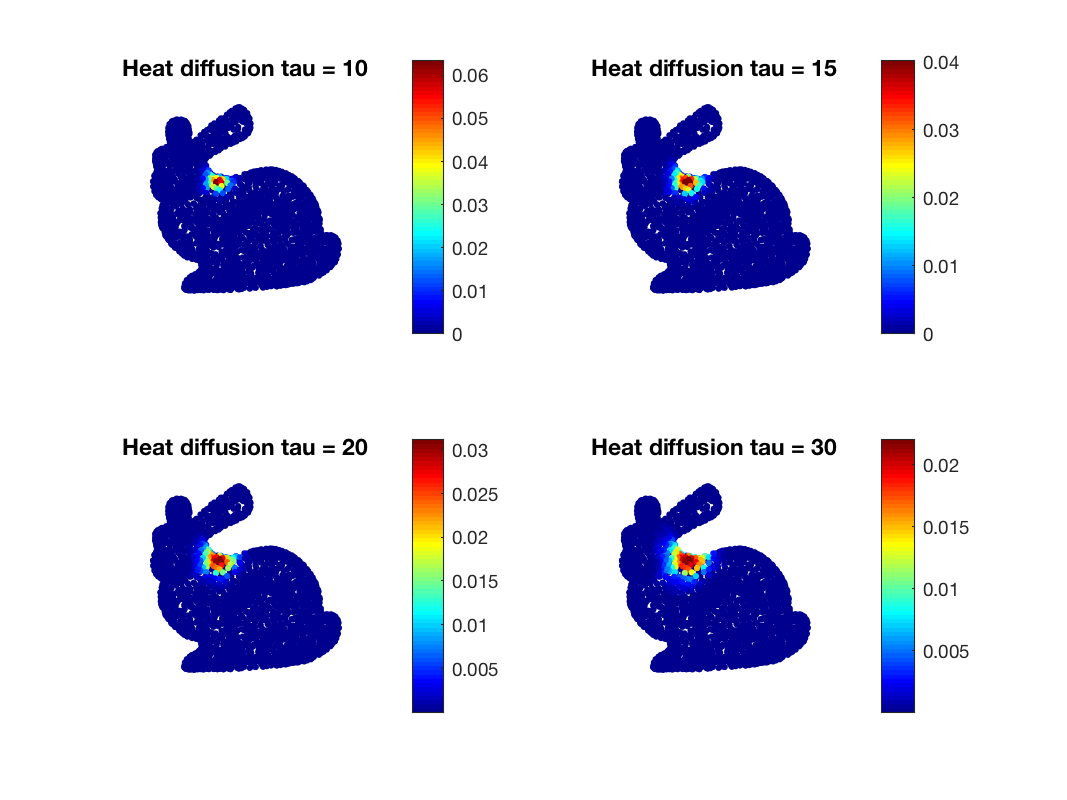
Heat diffusion at different scales
Visualizing wavelets atoms
Let's now replace the filtering by the heat kernel by a filter bank of wavelets. We can create a filter bank using one of the design functions such as gsp_design_mexican_hat
Nf = 6; Wk = gsp_design_mexican_hat(G, Nf);
We can plot the filter bank spectrum
figure; gsp_plot_filter(G,Wk);
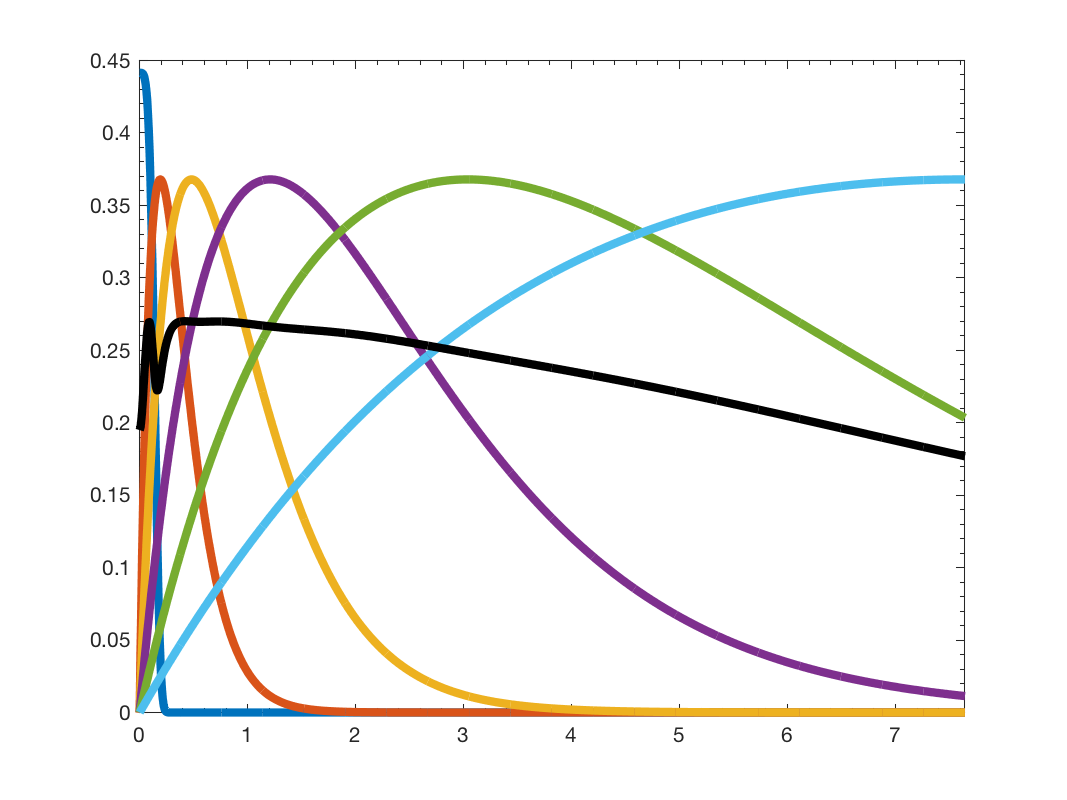
Wavelets filterbank (Original)
As we can see, the wavelets atoms are stacked on the low frequency part of the spectrum. If we want to get a better coverage of the graph spectrum we can use the function gsp_design_warped_translates
param_filter.filter = Wk; Wkw = gsp_design_warped_translates(G,Nf,param_filter);
Now let's plot the new filter bank
figure; gsp_plot_filter(G,Wkw);
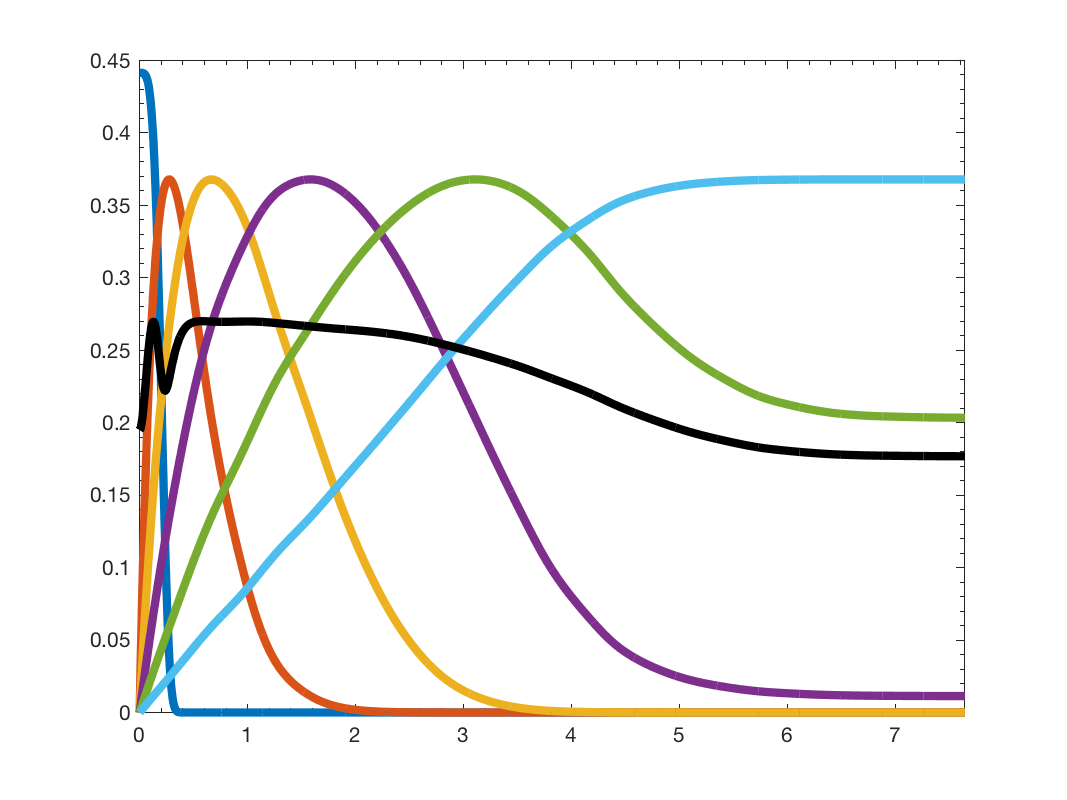
Wavelet filterbank (spectrum adaptated)
We can see that the wavelet atoms are much more spread along the graph spectrum. We can visualize the filtering by one atom as we did with the heat kernel, by placing a Kronecker delta at one specific vertex and filter using the filter bank
S = zeros(G.N*Nf,Nf);
S(vertex_delta) = 1;
for ii=1:Nf
S(vertex_delta+(ii-1)*G.N,ii) = 1;
end
Sf = gsp_filter_synthesis(G,Wkw,S);
We can plot the resulting signal for the different scales
figure;
subplot(221)
gsp_plot_signal(G,Sf(:,1), param_plot);
axis square
mu = mean(Sf(:,1));
sigma = std(Sf(:,1));
c_scale = 4;
caxis([mu - c_scale*sigma, mu + c_scale*sigma]);
title('Wavelet 1');
subplot(222)
gsp_plot_signal(G,Sf(:,2), param_plot);
axis square
mu = mean(Sf(:,2));
sigma = std(Sf(:,2));
caxis([mu - c_scale*sigma, mu + c_scale*sigma]);
title('Wavelet 2');
subplot(223)
gsp_plot_signal(G,Sf(:,3), param_plot);
axis square
mu = mean(Sf(:,3));
sigma = std(Sf(:,3));
caxis([mu - c_scale*sigma, mu + c_scale*sigma]);
title('Wavelet 3');
subplot(224)
gsp_plot_signal(G,Sf(:,4), param_plot);
axis square
mu = mean(Sf(:,4));
sigma = std(Sf(:,4));
caxis([mu - c_scale*sigma, mu + c_scale*sigma]);
title('Wavelet 4');
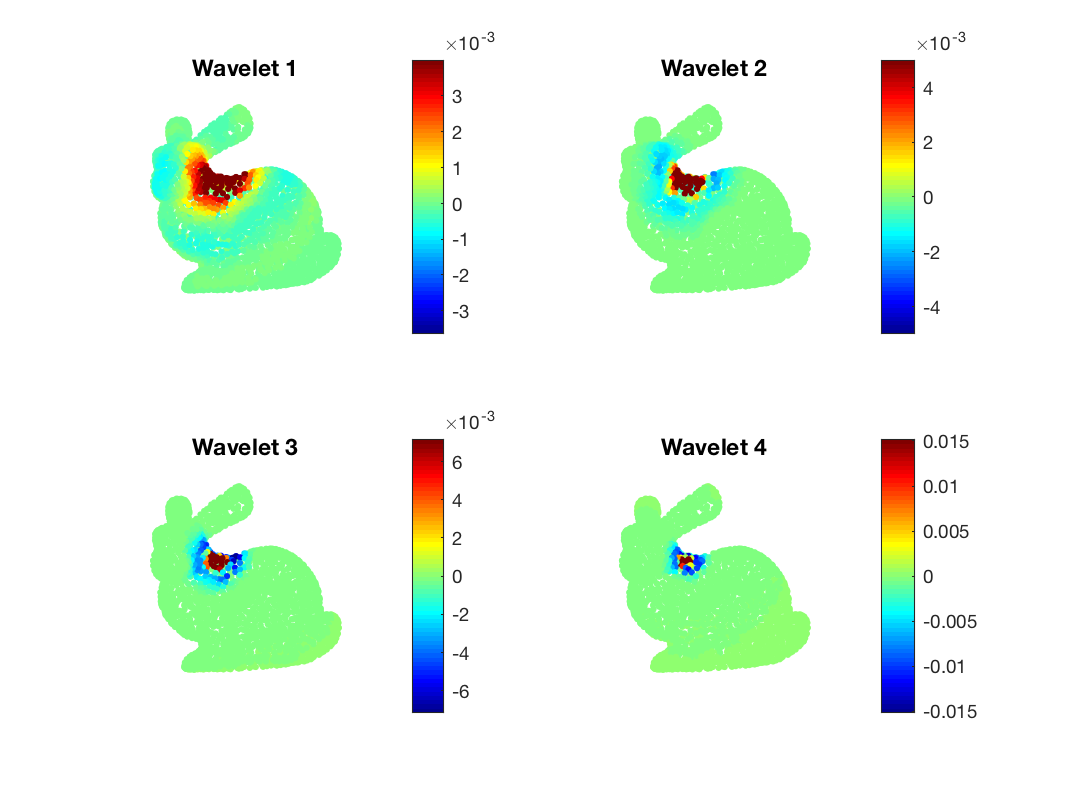
A few wavelets atoms
Curvature estimation
As a last and more applied example, let us try to estimate the curvature of the underlying 3D model by only using only spectral filtering on the graph.
A simple way to accomplish that is to use the coordinates map [x, y, z] and filter it using the wavelets defined above. We obtain a 3-dimensional signal [hat(x), hat(y), hat(z)] which describes variation along the 3 coordinates
s_map = G.coords; s_map_out = gsp_filter_analysis(G, Wk, s_map); s_map_out = gsp_vec2mat(s_map_out, Nf);
Finally we can get the curvature estimation by taking the l_1 or l_2 norm of the filtered signal
dd = s_map_out(:,:,1).^2 + s_map_out(:,:,2).^2 + s_map_out(:,:,3).^2; dd = sqrt(dd);
Let's now plot the result to observe that we indeed have a measure of the curvature
figure;
subplot(221)
gsp_plot_signal(G,dd(:,2), param_plot);
axis square
title('Curvature estimation scale 1');
subplot(222)
gsp_plot_signal(G,dd(:,3), param_plot);
axis square
title('Curvature estimation scale 2');
subplot(223)
gsp_plot_signal(G,dd(:,4), param_plot);
axis square
title('Curvature estimation scale 3');
subplot(224)
gsp_plot_signal(G,dd(:,5), param_plot);
axis square
title('Curvature estimation scale 4');
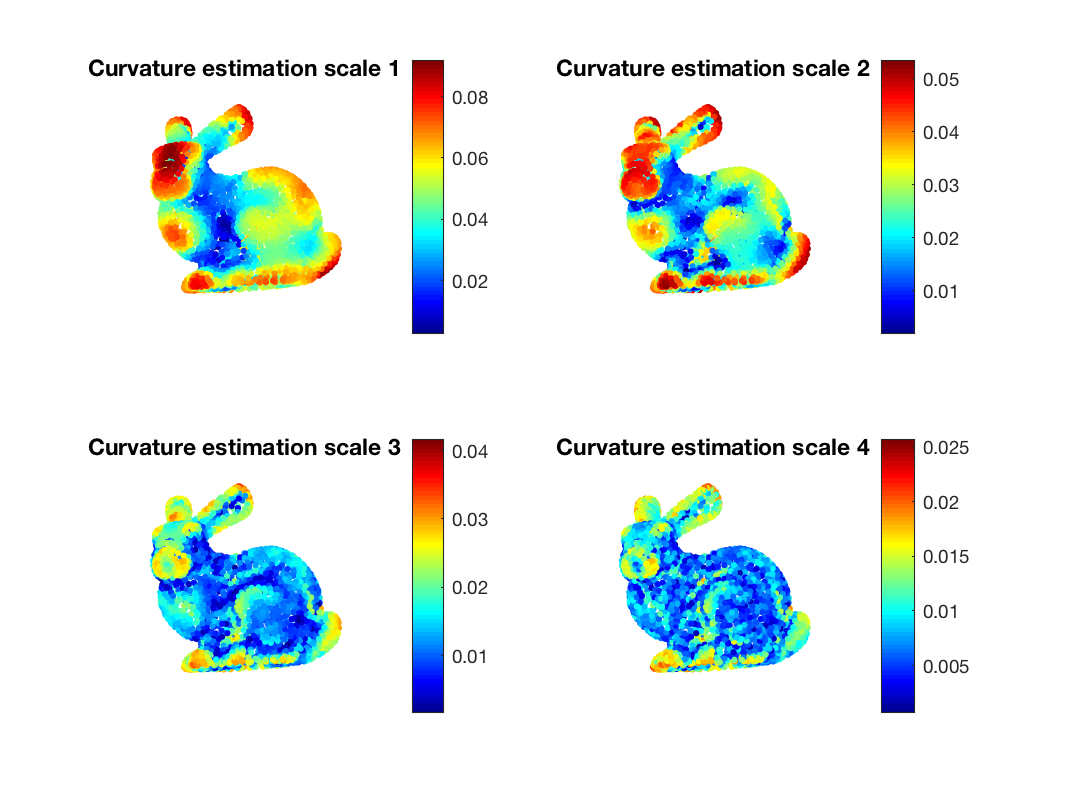
Curvature estimation using wavelet feature
This code produces the following output:
GSP_DESIGN_WARPED_TRANSLATES: has to compute the spectrum continuous density function approximation